THE BIG BAD BAC: BACTERIAL ARTIFICIAL CHROMOSOMES
(August 2004)
Genetic Carrying Handles: Cloning Vectors
In order to clone a gene, its DNA sequence must be attached to some kind of carrier, also made of DNA, that can take it into the cell. Biologists call these carriers vectors. A vector acts like a handle for the DNA, and it also contains other tools such as an origin of replication and a selective marker. The origin of replication is a sequence of DNA that the host cell recognizes that allows it to make more copies of the clone DNA sequence. This origin sequence is where the cell begins copying the vector and the attached clone DNA. The selective marker is a specific DNA sequence that is used by biologists to tell if the clone has entered the cell, and they are usually genes that confer antibiotic resistance to the cell. The most common media used for this process is actually very similar to chicken soup, but the carbohydrate agarose is added to convert the media into a semi-solid substance, since bacterial colonies are much easier to detect on a semi-solid surface. Agarose is much like gelatin, but it comes from seaweed and unlike gelatin, most bacteria cannot digest it. Antibiotics are often added to the media, to kill any cells that do not possess the antibiotic resistant selective marker gene, which is in the clone DNA. This way, biologists can ensure that all the remaining cells have in fact taken up the clone DNA and its vector. These cells are called transfected cells. Antibiotics are not the only way to identify transfected cells. Biologists sometimes use selective markers that turn cells a different colour or even to make them glow. Common proteins that do this include luciferase, which makes fireflies glow or green fluorescent protein, which comes from certain species of jellyfish. Green fluorescent protein also comes in other colours!
There are many different kinds of vectors and most of them are adopted from viruses or plasmids; small circles of DNA found in many bacteria and yeasts that are separate from their much larger genome. Phages are viruses that only infect bacteria, and some of them have been adapted into vectors. However, phages are sometimes hard to work with and they cannot contain much more genetic material than they already have wild-type.
Plasmids that have been turned into vectors can hold about 10,000 nucleotide base pairs (bp) or 10 kilobase pairs (kb). Cosmids are a weird mix between plasmids and phages and can hold up to 30 kb. In comparison, the average size of a human gene is about 27 kb. The average size of a gene in rice, an important and widely studied crop in many areas of the world, is about 2.3 kb. Of course, there are larger and smaller genes than these.
Viruses are another important area of study in molecular biology and they come in many different sizes. The genome of the herpes virus ranges from 100 kb to 250 kb, far bigger than the 10 kb that a plasmid vector can hold and beyond even the 30 kb carrying capacity of a cosmid. In addition, biologists are also trying to study sets of several genes that are adjacent to one another in a genomic sequence. The Human Genome Project read the sequence of all the genes in humans – that’s about 3 billion basepairs! Using plasmid vectors, more than 1.8 billion clones are required to examine an entire human genome.
Super-Sized Inserts
Bacterial Artificial Chromosomes (BAC) have been developed to hold much larger pieces of DNA than a plasmid can. BAC vectors were originally created from part of an unusual plasmid present in some bacteria called the F’ plasmid. The F’ plasmid allows bacteria to have “sex” (well, sort of: F’ helps bacteria give its genome to another bacteria but this only happens rarely when bacteria are under a lot of stress). F’ had been studied extensively and it was found that it could hold up to a million basepairs of DNA from another bacteria. Also, F’ has origins of replication and bacteria have a way to control how F’ is copied. In 1992, Hiroaki Shizuya took the parts of F’ that were important, cleaned it up, and turned it into a vector.
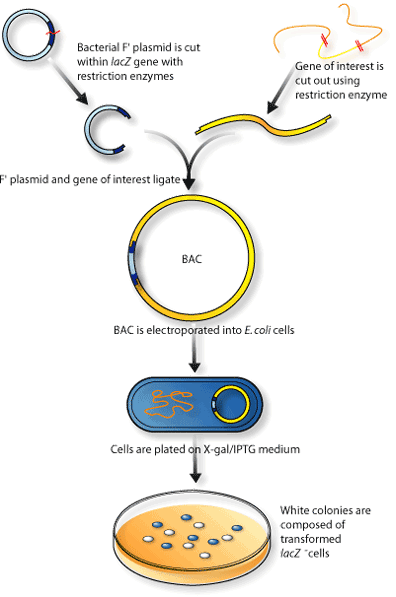
BAC vectors are able to hold up to 350 kb of DNA and have all of the tools that a vector needs to work properly, like replication origins, antibiotic resistance genes, and convenient places where clone DNA can insert itself. With these vectors it is possible to study larger genes, several genes at once, or entire viral genomes. By using a vector that can hold larger pieces of DNA, the number of clones required to cover the human genome six times theoretically could drop from 1.8 billion to about 50 million.
Researchers have modified BAC vectors to become more convenient to use and more useful in specialized situations. In addition to the antibiotic resistance gene that was added to identify transfected bacteria, a gene was added that enabled the bacteria to turn the colourless substance X-gal/IPTG blue. This substance is found in the chicken-soup-like media on which the bacteria colonizes. This colour-changing gene, called lacZ, is split apart when the clone DNA is incorporated into the vector, so it is possible to tell not only if a bacteria had been transfected (meaning incorporated into the cell), but also if the bacteria was transfected with the vector containing insert DNA or just the vector alone (remembering that if the vector has properly incorporated the clone DNA, it will have lost its ability to change X-gal/IPTG blue)(Figure 2).
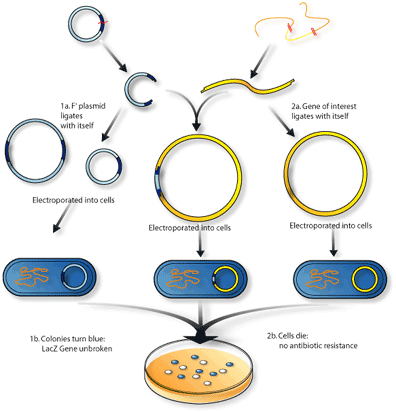
Another similar modification uses the gene called sacB, which encodes a protein called levansucrase. This protein turns sucrose – table sugar – into levan, a toxic substance to bacteria. In a similar fashion, bacteria grown on media with sucrose will die if sacB is not broken up by inserted DNA. If the vector carries a DNA insert, then sacB is broken and it won’t produce levansucrase, so the bacteria can survive in the sucrose media. In theory, only bacteria transfected with a vector containing insert DNA would be able to grow and form colonies.
Some modifications to BAC vectors make them more specialized. For instance, there are researchers studying the herpes virus who have made a BAC vector that can be cultured in bacterial cells and then when put into a mammalian cell, instantly releases its insert DNA – in this case, the whole herpes virus genome. With such a vector, it is easier to grow sufficient amounts of the herpes virus for research, since it can live in bacterial cultures, instead of requiring their endemic mammalian cell cultures, which are extremely difficult to maintain. This in turn makes it easier to make modifications to the virus and study what each of its genes do.
The creation of BAC vectors has allowed researchers to do many things that they could not do before and do them more quickly and more easily. Some of the scientific practices that have simplified with BAC vectors includes phylogenetic studies (studies which examine species’ relationship to one another) and what the absolute minimum size of a genome could be. The world is currently overrun by a plethora of microbial species that cannot be grown in cultures. BACs have allowed researchers to look at microbial DNA without having to actually grow the organisms, since the DNA is kept within easy-to-grow bacterial cultures. BAC vectors are also useful for studying pathogens, and are helpful in the development of vaccines. Many pathogens are becoming resistant to all antibiotics available to medicine and BAC vectors are playing a tremendous role in discovering new and powerful antibiotics in the environment. Discovering enzymes that are able to help clean up oil spills or help farmers breed healthier farm animals or even process radioactive waste are just a few examples of what Bacterial Artificial Chromosomes can do.
BAC vectors are useful tools and the methods for working with them are fairly well developed. Continuing advances such the modification of experimental animals will only increase the wide variety of uses for big bad BACs.
Glossary
Clone – a piece of DNA found inside a cloning vector
Genome – the entirety of an organism’s genetic material
Wild-type – organisms that have not been manipulated or changed by scientists
Pathogen – an organism or virus that causes disease
References
Shizuya, H., Birren, B., Kim, UJ., Valeria, M., Slepak, T., Tachiiri, Y., & Simon, M. 1992 Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. 89; 8794-8797 PNAS
(Art by Jen Philpot, slightly higher res version of second figure found here)