SYSTEMS BIOLOGY: AN OVERVIEW
Milestones in DNA research
The discovery of DNA structure in 1953 was the starting point of a real scientific and cultural revolution, the ending of which is difficult to predict. The discovery and use of enzymes that copy, cut and join DNA molecules in cells was the next step in this revolutionary course. The development of two major techniques contributed further to this already vertiginous process: the manual DNA sequencing method, which appeared in 1975, and the discovery of the polymerase chain reaction (PCR) in 1985, which allows the million-fold amplification of DNA sequences.
A natural consequence of these two breakthroughs combined with increasing computerized system capabilities was the automation of DNA sequencing. A prototype sequencing machine, able to sequence as many as 250 bases per day, appeared in 1986. By 1989, a robust instrument had been developed that could be used routinely in the laboratory. Fully automated and integrated sample preparation and sequencing were available in 1998. The latest sequencing machines are able to process over 1.5 million bases per day.
Another element that has played a key role in this revolution is the Human Genome Project (HGP) which officially started in 1990, although discussions and feasibility studies began earlier. The objective of the HGP was to generate a finished sequence in 15 years. Two versions of the draft of the human genome sequence were published in 2001. The HGP had a central role in this revolution two ways: it provided the driving force behind the development of high-throughput technologies (such as large-scale DNA sequencing, mass spectrometry and DNA arrays) and it constituted the practical application of the idea that the elements of a genome can be defined and catalogued for use in global analyses.
The underlying principle of this view of the human genome (or of any genome) is that it has a digital nature: it contains specific, clear information; it is a code. This information allows scientists to approach the study of all biological systems (they all share the same code) within a defined, fully delineated framework. The challenge is therefore to decode this information. In the opinion of some authors, Biology could be redefined as an essentially informational science. [1,2]
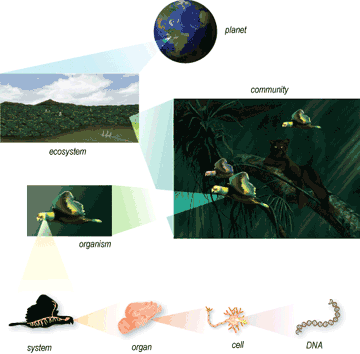
Digital nature of DNA and properties of biological systems
Biological systems contain two main types of digital information: genes, which encode the proteins through the intermediary of RNA, and regulatory networks which specify how these genes are expressed in time and space.
It is indeed remarkable that genetic information operates across such different time spans: Millions to tens of years (evolution and conservation of species), tens of years to hours (life cycles) and weeks to milliseconds (physiology) [1]. Moreover, biological information operates on multiple levels of organization (molecular, cellular, organic, systemic, etc.) and is processed in complex networks, which happen to be considerably robust (single perturbations will rarely cause systemic failure).[2]
The study of biological systems cannot be limited to simply listing its components (proteins, genes, cells, etc.); while an exhaustive list of all the parts of a car may give a vague impression, it does not necessarily help one to understand how the car functions. Similarly, a deeper understanding of biological systems can demonstrate how these parts are assembled together and how they interact with each other and with the surrounding environment. In other words, a system-level understanding is required. This is the objective of Systems Biology.
The systems approach brings with it a sense of wholeness. In words of Ludwig von Bertalanffy, the author of General System Theory, contemporary science should recognize the importance of “wholeness”. Wholeness is defined as “problems of organization, phenomena not resolvable into local events, dynamic interactions manifest in the difference of behavior of parts when isolated or in higher configuration, etc.; in short, ‘systems’ of various orders not understandable by investigation of their respective parts in isolation.” [3]
Systems Biology
The Systems Biology approach starts with the definition of the structure of the system under study (the structure of the overall network, be it composed of genes, proteins, metabolites, etc.). Then the attention shifts to the system dynamics, in order to determine its functional properties. These two aspects (structure and dynamics) provide a baseline that can be used to analyze an essential property of biological systems: robustness.
Robustness of biological systems manifests in various ways. Firstly, biological systems constantly adapt to internal or external changes. Secondly, they show certain insensitivity, which enables them to deal with the noise generated by the stochastic signals to which they are exposed. Finally, they also exhibit what could be called a graceful degradation, which is a slow and gradual end as opposed to the catastrophic failure that occurs when functions are damaged. [4]
The overall Systems Biology methodology includes the formulation of a model once the components of the system have been defined, followed by the systematical perturbation (either genetically or environmentally) and monitoring of the system. The experimentally observed responses are then reconciled with those predicted by the model. Finally, new perturbation experiments are designed and performed to distinguish between multiple or competing model hypotheses. [5]
The task that Systems Biology attempts to undertake is the actual integration of genomics, proteomics and indeed all the emerging omic disciplines, with the ultimate aim of designing biological systems.[4]
The omic information
So what are these omic disciplines? As the aim of genomics is to gain an insight into the entire genome of an organism, the genome being its entire set of genes, genomics is the study of the whole set of genes of a biological system. Likewise, the object of the study of proteomics is the proteome, understood as the entire collection of proteins that are expressed in a system. In this way, the respective discipline arises from the study of the transcriptome (the set of RNA transcripts of a specific system) and the metabolome (the entire range of metabolites taking part in a biological process). Other omes (sets) that may also be of interest include: the interactome (complete set of interactions between proteins or between these and other molecules), the localizome (localization of transcripts, proteins, etc.) or even the phenome (complete set of phenotypes) of a given organism. Systems biology strategies can thus be viewed as a combination of omic approaches, data integration, modeling and synthetic biology. [6,7]
Requirements for Systems Biology research
Evidently, the aims and approaches of systems biology are quite ambitious. It requires quantitative global High-Throughput (HT) biological tools such as DNA sequencing, DNA arrays, genotyping, proteomics, etc. Also needed are an extensive power of computational tools for databases and models and a very demanding integration of different levels of biological information, which requires the formation of cross-disciplinary scientists, not to mention the corresponding financial resources.
Conclusions
As previously mentioned, the discovery of the structure of DNA was a turning point in the history of science, culture and society. Its impact on medicine, agriculture, energy production, social issues, ethics, etc., continues to create interesting challenges in many areas of human activity. Scientists able to cross boundaries between many disciplines can make a valuable contribution to society. Awareness of the wholeness of this task as well as its implications, not only for science but for humanity, requires a sense of responsibility that is equally whole. [2]
References
1. Chong L et al., Whole-istic Biology, Science, Vol 295, no. 1, March 2002, p. 1661.
2. De Hoog C.L. and Mann M. Proteomics. Annu. Rev. Genomics Hum. Genet., 2004, 5: 267-293.
3. Ge H. et al. Integrating ‘omic’ information: a bridge between genomics and systems biology. Trends in Genetics, 2003, 19, 10: 551-560.
4. Hood L. and Galas D. The digital code of DNA. Nature, 2003, 421: 444-448.
5. Ideker T. et al. A new approach to decoding life: Systems Biology. Annu. Rev. Genomics Hum. Genet., 2001, 2: 343-372.
6. Ideker T. et al. Integrated Genomic and Proteomic Analyses of a Systematically Perturbed Metabolic Network. Science, 2001, 292: 929-934.
7. Kitano H. Systems Biology: A Brief Overview. Science, 2002, 295: 1662-1664.
(Art by Jen Philpot – note that high res versions of image file available here)