Reductive Genome Evolution of Obligate Symbiont Midichloria humanum: Implications of Gene Loss
(Click on the front page image to download the full article)
Abstract:
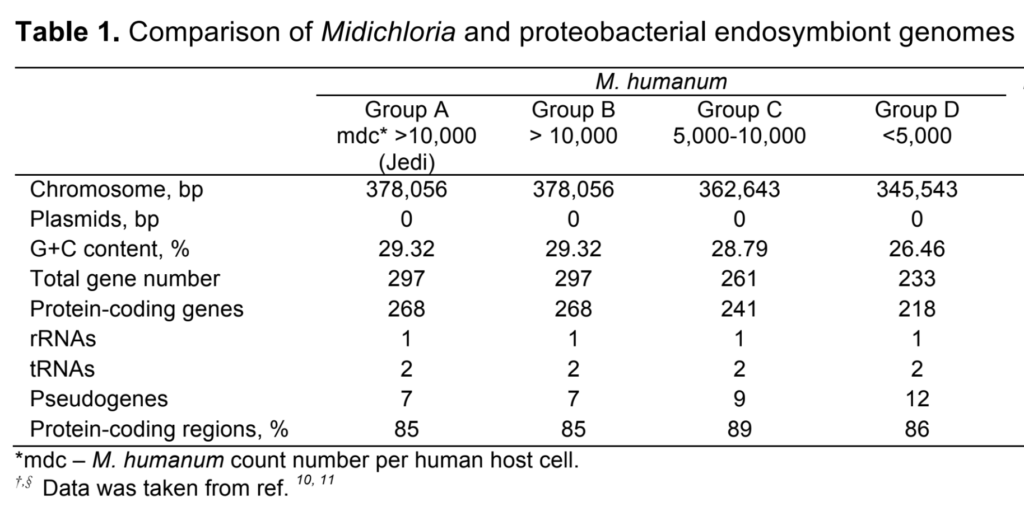
We sequenced and analyzed the genome of the Midichloria humanum, a ubiquitous intracellular symbiont, in 34 human subjects covering a range of population sizes from 1,000 to over 14,000 midichlorians per human cell. Midichlorians mediate the relationship between their human hosts and the Force, which confers exceptional physical and cognitive abilities on hosts with per-cell midichlorian populations (‘midichlorian count,’ mdc) larger than 10,000. Although, quantitatively, the genome of M. humanum is more extensively degraded than those of closely related Rickettsia species, midichlorians retained many intact genes involved in metabolic pathways, energy production and conversion, as well as nucleotide transport and metabolism. Wide-genome comparison between human groups classified based on mdc revealed that this symbiont continues to lose genes as a result of reductive genome evolution. Human groups with mdc < 10,000 have shown a loss of the DNA replication initiation gene dnaA that codes for the protein DNaA. Extensive polymorphisms, single nucleotide polymorphisms (SNPs) and deletion mutations have degraded the recA gene implicated in DNA recombination, which remains intact in the closely related species M. togrutas. A single mutation in the DNA repair gene mutY produced an inactive protein in groups with low mdc, further reducing pathways of DNA repair. The groups with medium and high mdc number retained active MutY protein. Overall, this work demonstrates that M. humanum is undergoing a slow genome reduction and that these genomic changes may be related to the continuous decline in birth rate of Force-sensitive children with mdc over 10,000.
Keywords:
midichloria, symbiont, genome, shotgun sequencing, gene loss, evolution.