CHANGING THE LANGUAGE OF DNA
DNA is the genetic code of life – a sort of molecular instruction manual that is passed on from mother to daughter cell. This set of instructions is read by the cell and translated into proteins, which perform specific functions within the cell. The DNA molecule itself is made up of a linear sequence of four deoxyribo-nucleotides: adenine (A), guanine (G), cysteine (C) and thymine (T), which in turn form the alphabet of genetic information. The sequence of this linear code leads to the synthesis of proteins through the cellular processes of transcription and translation. To give you a general picture of things, cells first transcribe the information from DNA into messenger RNA (mRNA), a sort of temporary copy of the gene. mRNA is then read three nucleotides at a time (known as a triplet codon – which if we consider nucleotides as letters of the alphabet, triplet codons are the words), translated into a string of amino acids, and, after a few modifications, becomes a protein. This series of events is commonly referred to as The Central Dogma. It’s how things work, rules to abide by if you like. However, recently, in the bacterium E. coli, scientists have altered the cellular machinery to read and translate a quadruplet codon[1], expanding the coding capability of DNA and the language of genes.
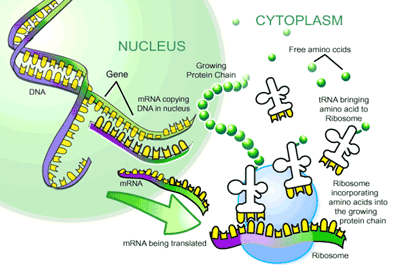
mRNA to Protein: the Translational Machinery
Translation is the process where mRNA is read and translated into a string of amino acids. This process takes place at organelles called ribosomes, which bind and slide along the mRNA and serve as a framework for translating the genetic message. As each triplet codon is read (for example AGC), a transfer RNA (tRNA) molecule brings a specific amino acid to the ribosome, and this amino acid is then chemically joined to the previous amino acid by a peptide bond. Basically, these tRNA molecules are like waiters. Each is trained to take a specific order from a certain triplet codon and fetch the amino acid corresponding to that order. On one arm of the ‘T’ is an anticodon loop containing the complimentary triplet codon (for ACG, it would be UGC) to that on the mRNA. On another arm is an acceptor stem that attaches to the amino acid corresponding to the triplet codon (in this case serine). After the tRNA molecule delivers its amino acid to the translation complex, it floats away to be recharged with another amino acid. This is done by enzymes called aminoacyl-tRNA synthetases (ARS), which recognize both the anticodon loop and acceptor stem of tRNAs and attach the corresponding amino acid. These ARSs essentially ensure that the tRNA waiters pick up the right order. After the string of amino acids is transcribed, it is folded and modified to become a protein.
Unnatural Amino Acids and Protein Study
So why would anyone want to alter the language of genetic information? The answer lies with our need to study proteins and the cellular processes for which they are responsible. An expanded genetic code allows for the incorporation of unnatural amino acids into proteins, opening the door to previously unavailable experimental methods. Although mutating DNA through site-directed mutagenesis has long been used to exchange one natural amino acid for another, this approach is limited to exchange between properties found in nature. For instance, the amino acid asparagine, which is polar, can be exchanged for leucine, which is not. Unnatural amino acids, however, can be created with properties that can serve as novel tools in protein study [2]. Here are a few possibilities:
– Biophysical Probes: Modified amino acids containing fluorescent groups can be used to visualize protein localization and protein-protein interactions within a cell. Amino acids with synthetically added modifications, like phosphorylated or glycosylated side chains, may also be used to study the effect of these natural intracellular modifications on protein structure and function.
– Photoreactive Groups: Amino acids with photoreactive groups can be used as a light-induced trigger that can activate a certain protein function. Studies have used ‘caged’ sidechains that hide reactive or important parts of a protein. Photodecaging then serves as a way to ‘turn on’ the system for study.
– Stuctural Labels: Heavy atoms or spin labels can be used in methods used for structural studies such as spectroscopy, NMR, and crystallography.
Previous Work in Genetic Code Expansion
At the end of every gene is one of several stop codons. These lead to the recruitment of a corresponding tRNA that ends transcription of the gene. The discovery that certain mutated tRNAs could suppress these amber or stop codons was made thirty years ago [3]. Since then, amber suppressor tRNAs have been extensively studied as a means to insert an unnatural amino acid into a protein.
There is, however, a problem with trying to use stop codons to code for unnatural amino acids; there are only so many of these stop codons. Consequently, such a modified translational system may at best add one or two modified amino acids into a protein. Having suppressor tRNAs encoding stop codons could also affect the translation of numerous other genes. The use of an expanded genetic code with a quadruplet codon as a means of adding unnatural amino acids by-passes these restrictions. All modified tRNAs and aminoacyl-tRNA synthetases can use an entirely new set of four-letter genetic “words” independent of the natural triplet codons.
tRNAs able to read a quadruplet codon have been found naturally as frameshift suppressors. They have been so named because they suppress mutations in DNA that add a single nucleotide. Such an addition would, without the presence of the frameshift suppressor, cause all subsequent triplet codons to be out of phase. In other words, if a single letter is added to the sentence “THE CAT WAS FAT” and the spacing is kept the same, the sentence becomes “THX ECA TWA SFA”. The sentence no longer makes sense. The same thing happens to genetic codes when you add an extra “X” nucleotide, this is called a frameshift mutation. Analysis of these frameshift suppressor tRNAs revealed that they had a single added base in their anticodon loop, allowing them to read a corresponding four base codon on mRNA. Since their discovery, modified tRNAs have been used to incorporate natural amino acids in response to quadruplet codons in E. coli [4]
Quadruplet Codons and Unnatural Amino Acids
In order to create an in vivo system capable of transcribing a quadruplet codon to an unnatural amino acid, you need four things. (1) The first requirement is an unnatural amino acid. In their paper, Schultz et al. used homoglutamine since it is very similar to the naturally-occurring glutamine residue. (2) A mutated gene containing an extra nucleotide making a quadruplet codon is also needed. In this case AGGA was used. This is relatively easy to create using the technique of site-directed mutagenesis. (3) Also required is a mutated tRNA that can be specifically charged with the unnatural amino acid and that is able to recognize the quadruplet codon. (4) Lastly, a mutated aminoacyl-tRNA synthetase is needed to specifically charge the aforementioned tRNA with the unnatural amino acid. These last two components occupied the most time in the Schultz lab.
For the experiment to work, both the quadruplet codon tRNA and aminoacyl-tRNA synthetase must be orthogonal. This means that the tRNA cannot be charged with natural E. coli aminoacyl-tRNA synthetases and the homoglutamine-tRNA synthetase must not charge the natural E. coli tRNAs. This ensures that only the gene with the added quadruplet codon can incorporate homoglutamine into the protein. To increase the likelihood of orthogonality, the candidate tRNA and ARS were chosen from members of the archaebacterium family – chosen because they are very distinct from the E. coli versions. For the tRNA to read the AGGA quadruplet codon, the sequence of the anticodon loop was changed to the complimentary UCCU sequence. In addition, the acceptor stem was mutated to accommodate attachment of the unnatural homoglutamine amino acid.
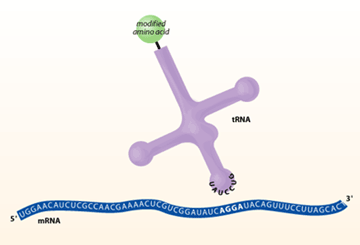
to a protein using a quadruplet codon.
Ensuring that the aminoacyl-tRNA synthetases (ARS) charged the mutated tRNAUCCU with homoglutamine required two changes. First, specific residues in the ARS active site were changed so it would bind both the tRNAUCCU acceptor stem and homoglutamine. Second, the part of the ARS that recognized the tRNA anticodon loop was deleted. This saved having to change to ARS to recognize a quadruplet codon on the tRNA. Since recognition of the proper tRNA already occurs in the active site, specificity is maintained.
Amazingly, with these few changes to two key players in the transcriptional process, Shultz et al. changed a fundamental process of life. This preliminary work demonstrates that the possibility of modifying genes to better understand their function is not limited to the natural amino acids and standard triplet codon. Although in this case a close homolog was used as the unnatural amino acid coded for, there is potential for utilizing novel residues to investigate protein structure and function.
References
Anderson, J.C., Wu, N., Santoro, S.W., Lakshman, V., King, D.S., & Schultz, P.G. An expanded genetic code with a functional quadruplet codon. PNAS 101, 7566-71 (2004).
Dougherty, D.A. Unnatural amino acids as probes of protein structure and function. Current Opinion in Chemical Biology 4, 645-52 (2000).
Capecchi, M.R., Hughes, S.H., Wahl, G.M. Yeast super-suppressors are altered tRNAs capable of translating a nonsense codon in vitro. Cell 6, 269-77 (1975).
O’Connor, M. Insertions in the anticodon loop of tRNAGln (sufG) and tRNALys promote quadruplet decoding of CAAA. Nucleic Acids Research 30, 1985-90 (2002).
(images by Jane Wang and Jen Philpot)