A MONK’S FLOURISHING GARDEN: THE BASICS OF MOLECULAR BIOLOGY EXPLAINED
(August, 2003)
The first inklings of genetic theory can be traced back to a common human experience: the recognition that a child has features similar to those of its parents. This ancient observation is actually one of the cornerstones of genetics and its subsequent offspring, molecular biology.
For centuries there was little evidence beyond the anecdotal that transmitted inheritance was a reasonable theory. Though it seemed sensible that a child with the same appearance as its parents likely received these characteristics from them, little evidence supported the notion and instead a good deal of confusion surrounded it. Part of the confusion arose from years of traditional breeding programs that sought to improve the quality of domestic plants and animals. The results were often highly unpredictable, with traits like sterility and disease susceptibility arising in the offspring, apparently from nowhere. Where were these characteristics coming from?
The beginnings of an answer first appeared in a monastery garden located in Brno, Czechoslovakia. It was there that an Augustinian monk named Gregor Mendel (1822-1884) had placed a pea plant, Pisum sativum, with the intention of carrying out a painstakingly long breeding experiment1 (see Figure 1).
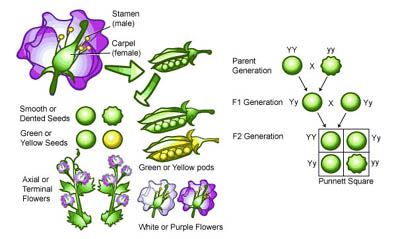
After analyzing 21,000 hybrid plants, Mendel conceived of the idea that individual units of inheritance existed, are discreet, and that two such units (one from the female parent and one form the male parent) combined to produce a characteristic of an offspring [1]. The concept of one unit of inheritance, later to be called a gene, was born.
Modern molecular biology has flowed from Mendel’s concept of transmissible genes. It was a starting point that led biologists to the identification of DNA as the primary genetic material, the uncovering of the biochemical structure of genes, an understanding of how DNA stores and regulates the flow of genetic material, and ultimately the development of techniques that allow for the manipulation of DNA.
Hunting for the Molecular Nature of Elusive Genes
While Mendel grew peas in his garden (around the year 1866) many biologists were focused instead on the use of a microscope to document the appearance of the smallest components of a living organism, the cell. A large variety of cells from different organisms were examined and in each case a similar morphological region called the nucleus was seen. Interestingly, certain dyes were found to stain small discreet bodies in all the different nuclei. These small bodies became known as chromosomes (meaning ‘coloured body’). By the turn of the century the insights Mendel had provided concerning transmissible units of inheritance began to be appreciated, and a fascinating possibility arose: could genes be located on the chromosomes that resided in the nucleus of a cell and somehow be transmitted to the next generation?
A few eyebrows were raised by the question, as it suddenly seemed possible that Mendel’s transmissible genes were actually cellular structures, which meant that they were both physically identifiable and could be subject to experimentation. Understandably, excitement about the subject grew. The wait for further breakthroughs was not long, as discoveries began to trickle out of Columbia University (USA) between 1905-1915. There, careful microscopic observation detected chromosomal differences between the sexes: the presence of two X chromosomes in cells from a female, and one X chromosome and one smaller chromosome shaped like a Y in the cells of a male [2]. Not only was it found that these chromosomes determined sex (XX = female, XY = male), but it was also shown that certain traits (and thus their genes) were transmitted only with the X chromosome [3] (see Figure 2).
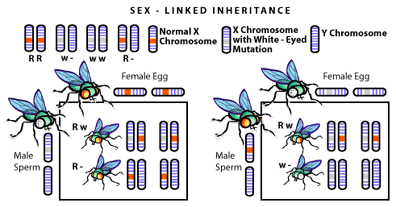
With chromosomes implicated in carrying the genes responsible for inheritance the question crossing every scientists mind was what were chromosomes made of?
It was here that a turning point was achieved. As biologists turned to the nucleus, trying to define the molecular nature of the chromosome, a shift in the field of biology occurred. In the years to come, the molecular biologish would take centre stage as the hunt for the molecular structure of a gene continued.
Methods available at the time made it very difficult to obtain a pure preparation of chromosomes, they were always contaminated with other cellular components. Nevertheless, it was discovered that chromosomes contained two components: (1) deoxyribonucleic acid, or DNA (commonly abbreviated to nucleic acid), and (2) basic proteins called histones. Even though DNA was present in much higher quantities than protein in these preparations, it was hotly debated whether or not the DNA or histones carried the genes biologists were looking for. A crucial point that kept biological circles divided was the relative structural simplicity of DNA, which was made up of four building blocks, as compared to the complexity of proteins, which were made up of 20 building blocks. Scientific opinion differed on what kind of structural complexity genes would require to dictate the intricacies of a cell; thus whether the cell used a “genetic protein” or “genetic DNA.” It took significant effort to resolve this debate, but in 1952 (many years after the 1915 studies on chromosomes) Alfred Hershey and Martha Chase [4] were able to use different radioisotopes to label proteins (35S) and DNA (32P). This technique allowed them to reveal that bacterial viruses, which were composed only of protein and DNA, reproduced themselves within bacteria by using only their DNA component. Thus, the debate was resolved: genes were made of DNA.
The Biochemical Structure of DNA is Unraveled
While the debate fumed over “genetic protein” versus “genetic DNA” in the 1920s, much about the chemical nature of nucleic acid was elucidated [5]. It was found to be composed of regularly repeating subunits called nucleotides. Only a limited number of nucleotides were found to exist in nature and all contained three elements: (i) a phosphate group(s) linked to a (ii) sugar, which was joined to a (iii) flat ring molecule commonly called a base (see Figure 3). The limited number of natural nucleotides is partially a result of the fact that only five types of natural bases exist: guanine (G), adenine (A), cytosine (C), thymine (T), and uracil (U). Each nucleotide was found to possess the ability to link to others to form chains. Surprisingly, only two similar types of chains existed, DNA and RNA. The most obvious difference between the two types was that the base uracil was only found in RNA, while the base thymine was found only in DNA.
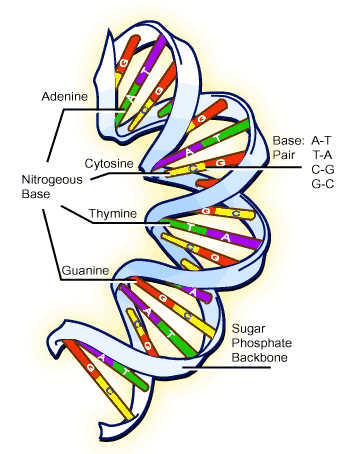
With the relatively simple chemical composition of DNA understood, a more philosophical question still remained: how did DNA govern and dictate the natural variety of life on Earth? This question boiled down to a crucial missing link, how the chemical structure of DNA, essentially a chain of nucleotides linked together, enabled it to act as a carrier of inheritance.
As this question was being asked, another interesting fact was obtained about the chemical characteristics of nucleotides; their bases (G, A, C, T, U) could chemically bind to each other. Not only that, they did so in an exceptionally specific manner. Adenine bound only to thymine in DNA (and uracil in RNA), while guanine bound only cytosine. As a consequence of this the amount of adenine equaled the amount of thymine, while the amount of guanine equaled the amount of cytosine in a DNA molecule [5]. This was later known as Chargaff’s rule, after the Austrian chemist Erwin Chargaff.
Soon after this was discovered, X-ray diffraction studies showed that DNA adopted a regular and precise helical structure. Enough data was now in place for a famous leap of scientific faith to be taken. In 1953, Watson and Crick [6] correctly deduced that DNA forms a double helix with two strands of nucleotides wrapped around each other (see Figure 3). The binding rules for nucleotides ensured that each strand was a complementary copy of the other (for example an adenine in one strand was always bound to thymine in the other strand). Thus, the two strands were complementary anti-parallel chains of nucleotides wound around each other to form a double helix. Our understanding of the molecular nature of inheritance took a step forward with Watson and Crick’s leap of logic, for it was immediately understood that such a structure would provide DNA with a simple mechanism to accurately reproduce itself: just pull the two strands apart and use one strand to create a complementary copy of the other using the nucleotide binding rules [7]. If done for both strands, two exact copies of the original DNA molecule would be created, a process eventually shown to be exactly the way DNA is copied in a cell.
The realization that DNA formed a double helix solved a large part of the question of how DNA was involved in all lifeforms by revealing how to make endless copies of a DNA molecule. But it would still be years before the molecular mechanism of inheritance was fully understood.
Cracking the Genetic Code
At the turn of the 20th century many scientists were beginning to turn their attention to the biochemical basis of heredity. Investigators in many disciplines wanted to understand the underlying biochemistry that dictated the physical appearance of an organism. The study of proteins took center stage. Of particular interest was an important class of proteins, termed enzymes. These proteins were able to catalyze biochemical reactions and were soon found to be responsible for biochemical function. During this period it was determined that proteins were composed of 20 naturally occurring amino acids linked together in a chain (called a polypeptide), much like DNA.
Even before the composition of genes was known, a link between proteins and genes was evident from the study of diseases in which cells fail to perform known biochemical reactions. An example was alkaptonuria, a rare genetic disease resulting from a failure to correctly breakdown two amino acids (phenylalanine and tyrosine) found in a regular diet. The build up of by-products from this blocked pathway produces the black urine characteristic of a patient with the disease. In 1908, Archibold Garrod [8] correctly surmised that the absence or deficiency of a given enzyme required for normal cellular biochemistry, in the case of alkaptonuria it was an absence of an enzyme required to break down amino acids, resulted in a metabolic disease. Since such defects in proteins could be inherited, it appeared that genes could dictate the production of the proteins in an organism. It took almost three decades for adequate knowledge to be gained about cellular metabolism to determine if Archibold Garrod’s link between genes and proteins was true. In the end, it was once again studies of defects in well-known metabolic reactions that showed that a gene directed the production of a single protein, a fact which is now generalized as the “one gene = one protein” rule.
The “one gene = one protein” rule and the understanding that genetic information is specified in the four nucleotide bases of DNA (A, C, G and T) led to a period of scientific excitement in which scientists wondered how four bases of DNA could encode the 20 known amino acids that make up proteins? By the 1950’s, scientists simply assumed the linear sequence of nucleotides in a DNA strand corresponded to the linear sequence of amino acids in a protein polypeptide. The first biochemical evidence of this assumption was that the position of mutations in a protein correlated to the position of mutations is a gene (i.e. they appeared in the same relative places in the molecules). A co-linear relationship seemed to exist between the two. A quick mathematical calculation determined that at least three nucleotides would be required to specify each of the 20 natural amino acids, since blocks of two nucleotides could only be combined to code for 16 of the 20 known amino acids and single nucleotides could only code for four. Random combination of four nucleotides produces 64 possible triplets, but it was not clear at the time how these 64 combinations would code for 20 amino acids.
In 1961, a historic set of experiments were begun by Marshall Nirenberg and Heinrich Matthaei [9] that solved the above question and marked the beginning of modern molecular biological techniques. They were able to create a synthetic nucleotide chain composed only of uracil (i.e. UUUUU), which they went on to add to cells that had been broken apart. When they did this they witnessed the production of a polypeptide chain. Even more interesting, it was composed of only a single amino acid, phenylalanine. Next, they began adding defined lengths of uracil chains to the extracts. By doing this they found that only multiplies of three nucleotides produced amino acid chains. For example, a chain of three uracils (UUU) gave a single phenylalanine, similarly a chain of four or five uracils (UUU-U or UUU-UU) also produced only one phenylalanine, while a chain of six (UUU-UUU) produced two phenylalanines linked together. Hence it was determined that a triplet of uracils in a gene coded for the amino acid phenylalanine in a protein. Production of all the possible combinations of three nucleotides (called a codon) soon revealed which triplets coded for which amino acids. It was found that 61 combinations coded for the 20 amino acids, while the three remaining codons were used as “stop” signals for the end of a protein.
The genetic code was now broken. Scientists understood how a protein was encoded in the molecular structure of DNA. With this information it was not long before the underlying mechanisms of how a cell used DNA to make protein became clear.
Producing Genetic Messages from DNA
All living organisms depend upon the production of proteins encoded by the information held within their DNA. Despite the variations that exist between organisms, it was soon found that all cells make use of the same general mechanism for decoding the information in DNA into proteins, termed gene expression. Even though the amino acid sequence of proteins is dictated by the nucleotide sequence of many genes, proteins are not directly synthesized from DNA. Instead, genes produce proteins in two discreet stages, which involve many different types of enzymes, proteins and RNA molecules.
The first stage is called transcription (see Figure 4), in which an RNA copy (or transcript) of a specific gene is produced. This RNA copy of the gene is called a messenger RNA (mRNA), since it is the genetic message that will produce a protein. Production of mRNA requires an enzyme called RNA polymerase. It begins the process by binding to specific nucleotide sequences in the DNA (called a promoter), located just up from the gene that specifies a protein. A complex process unwinds the DNA in this area so that the polymerase can begin to move along the DNA strand like a train along a rail. As the RNA polymerase moves along it synthesizes an RNA copy according to the nucleotide sequence it encounters (done by pairing a new nucleotide to a complementary nucleotide in the DNA using the base pairing rules, followed by linking them together). This procedure continues until the polymerase hits a defined sequence called a terminator, which causes the polymerase to fall off the gene and release the mRNA. Once released, an mRNA is free to float through the cell bearing its genetic message and ultimately engages in the second stage of producing a protein from a gene.
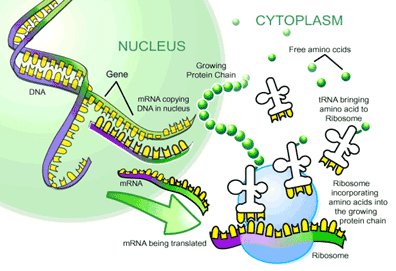
ecoding Genetic Messages
The process of decoding mRNA transcripts is termed translation and once again involves many types of proteins and RNA molecules. In particular, translation requires two types of RNA termed ribosomal RNA (rRNA) and transfer RNA (tRNA). Ribosomal RNA is intimately involved in the synthesis of proteins through the interaction of various types of rRNA to form a complex called a ribosome. This is the cellular machine that creates proteins from mRNA. A ribosome forms a donut structure with the mRNA passing through its center; a specific site within the mRNA (called the ribosome binding site or RBS) then binds to the ribosome, causing the second type of RNA, the tRNA, to spring into action. tRNAs are universal adaptor molecules that carry amino acids and a complementary triplet of nucleotides called an anti-codon that recognizes each codon in an mRNA. As mentioned before, codon is the name given to triplets of nucleotides in mRNA that code for particular amino acids. An anti-codon is just a complement of a codon. Through this method each triplet in an mRNA molecule will bind to a tRNA that bears its complementary triplet codon. For example, a string of A-U-G nucleotides in an mRNA will bind a tRNA that has a U-A-C triplet, all of which is based on the nucleotide binding rules. Since each tRNA molecule carries an amino acid, the triplet codon will result in a specific amino acid being brought to the ribosome. At the ribosome these amino acids are bound together into a polypeptide chain (i.e. a protein) in exactly the linear order that the mRNA dictates, a sequential process that will produce a protein corresponding directly to the nucleotides in the mRNA.
Central Dogma of Molecular Biology
Over a century of work has gone into the current framework of molecular biology, but at its core, our understanding can be broken down to what has become known as “the central dogma” of molecular biology: DNA is copied into genetic messages, which are then translated into proteins that go on to perform the underlying biochemistry of an organism.
The hunt to understand inheritance has been an important scientific journey. It has required a broad range of disciplines, from chemistry to microbiology to zoology, as well as the development of many new and exciting technologies. This integration of disciplines, combined with the resolution of many engineering difficulties, eventually resulted in the emergence of the field of biotechnology,an exciting enterprise that is bringing molecular biology to the forefront of society and causing us to reshape the way we see the world.
Additional Reading and Texts Consulted
1. Alberts B. et al, ed. (2002). Molecular Biology of the Cell. New York/London: Garland Publishing. 1616p.
2. Snyder L., Champness W., eds. (1997). Molecular Genetics of Bacteria. Washington DC: ASM Press. 504p.
3. Waston J.D. (1968). The double helix: a personal account of the discovery of the structure of DNA. New York: Atheneum. 226p.
4. Singleton P., Sainsbury D. (2001). Dictionary of Microbiology and Molecular Biology. New York/West Sussex: Wiley. 895p.
5. Clark D.P., Russell L.D. (1997). Molecular Biology: made simple and fun. Vienna: Cache River Press. 470p.
5. Raineri D. (2001). Introduction to Molecular Biology. Malden MA: Blackwell Science. 190p.
References
1. Peter JA, ed. 1959. Mendel’s experiments in plant hybridizations. English translation reprinted in: Classic Papers in Genetics. New Jersey: Prentice-Hall.
2. Sutton W.S. The chromosome in heredity. Biol. Bull. 4: 231-251.
3. Morgan T.H. (1910). Sex-limited inheritance in Drospohila. Science 32: 120-122.
4. Hershey A.D., Chase M. (1952). Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36: 39-56.
5. Chargaff E. (1951). Structure and function of nucleic acids as cell constituents. Fed. Proc. 10: 654-659.
6. Watson J.D., Crick F.H.C. (1953). Molecular structure of nucleic acids: a structure for deoxyribonucleic acid. Nature 171: 737-738.
7. Watson J.D., Crick F.H.C. (1953). Genetical implications of the structure of deoxyribonucleic acid. Nature 171: 964-967.
8. Garrod A.E. (1908). Inborn errors of metabolism. Lancet 2: 1-7,73-79,142-148,214-220.
(Art by Jane Wang – note that high res versions of image files available here)