SPOT YOUR GENES – AN OVERVIEW OF THE MICROARRAY
(August 2004)
A cell functions by using its genes to produce proteins and although each cell within an organism will usually contain the same set of genes, there are significant differences in which genes are activated and how they are controlled. This is idea is easy to digest when you think about how a single-celled embryo goes on to produce all kinds of different tissues. At any rate, the mechanism by which genes are utilized is the same for all cells and involves the transcription of a gene into mRNA before being translated into a protein. The production of mRNA is very much a reflection of the activity level of a gene, and a lot of genetic information can be understood by studying it. For many years, study of a gene in this manner had to be done individually—looking at whether a specific gene is turned on (upregulated) or turned off (downregulated) under certain conditions1. Enter the microarray: a technological advancement that within the last 7 or 8 years has allowed scientists to study many, if not all, the genes of an organism’s at once [1]. This high throughput method allows for the global study of changes in gene expression, giving us a complete cellular snapshot.
Introducing…a DNA Microarray
A DNA microarray consists of an orderly arrangement of DNA fragments representing the genes of an organism [2]. Each DNA fragment representing a gene is assigned a specific location on the array, usually a glass slide, and then microscopically spotted (less than 1 mm) to that location. Through the use of highly accurate robotic spotters, over 30,000 spots can be placed on one slide [2], allowing molecular biologists to analyze virtually every gene present in a genome. The main advantage of microarrays is that the spots are single stranded DNA fragments that are strongly attached to the slide, allowing cellular DNA or RNA to be fluorescently labeled and laid overtop of the array. DNA or RNA in the overlaid sample will stick (through a process called hybridization) to a complementary spot on the array, that is, Gene-A will stick to a spot composed of a Gene-A fragment. By exposing the microarray to a fluorescently labeled sample the DNA that hybridizes will be identifiable as glowing spots on the array, while the spots that have nothing hybridized to them will not be visible. Two main types of commercial microarrays, oligonucleotide arrays (Affymetrix) and cDNA arrays (BD Biosciences and others), provide good examples of how this technology works.
Oligonucleotide Arrays
Oligonucleotide arrays (trademarked as a GeneChip by Affymetrix) use small 25 base pair gene fragments as the DNA to be spotted onto an array [3]. In all, 11 to 16 copies of this DNA are spotted for each gene to be placed on the array. These DNA fragments, often called probes, are selected to have little cross-reactivity with other genes so that non-specific hybridization will be minimized. Nevertheless, some non-specific hybridization will occur; to combat this, a second probe that is identical to the first except for a mismatched base at its centre is placed next to the first. This is called the Perfect Match/Mismatch (PM/MM) probe strategy. Any background hybridization with the MM probe is subtracted from the PM probe signal which results in perfect hybridization.
Oligonucleotide arrays are printed using a combination of photolithography and combinatorial chemistry [4] (see Figure 1), a technique that allows the simultaneous generation of thousands of probes relatively quickly. Their production begins with a 5-inch square of quartz wafer, it being an ideal substance on which chemicals adhere.
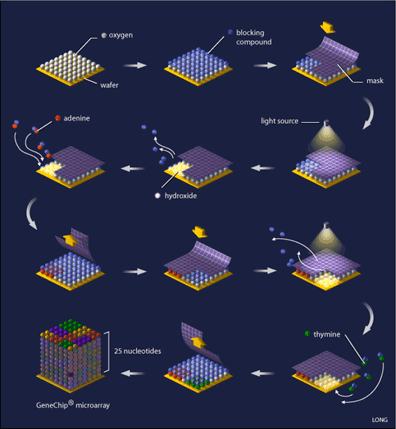
This wafer is washed with a blocking compound that can be removed by exposure to light. What is needed next is referred to as a ‘mask’, (see Figure 1) which is designed with 18-20 micron square windows that allow light to pass through areas where a specific nucleotide is needed, for example, if the mask is designed for the addition of adenine to the probe, you will find a window at that location, while probe locations where another base is required are blocked. Such a mask is laid on top of the array and ultraviolet light is shone onto it. Probe locations that have windows are exposed to the light and the blocking compound removed from that probe, while covered locations still have the blocking compound present. The quartz wafer is then washed with a solution of the desired nucleotide (eg. adenine) that is linked to the same blocking compound, causing the nucleotide to attach to the probes that were exposed to light, while the nucleotide-attached blocking compound ensures that all of the probes are protected again. A capping step is added so that probes that did not attach their appropriate nucleotide are not incorrectly synthesized. The array is now ready for a new mask and the addition of a new nucleotide, a process that is repeated until all of the probes are complete (25 nucleotides each). Computer algorithms can calculate the optimal design of the masks that will minimize the number of reactions needed.
Once constructed, an oligonucleotide array is ready for use in the laboratory. It is generally used for one of two things: (1) quantification of the amount of mRNA in a single sample (e.g. to determine the amount of mutated vs. non-mutated mRNA) or (2) the comparison of two different samples hybridized to two separate arrays [3-4].
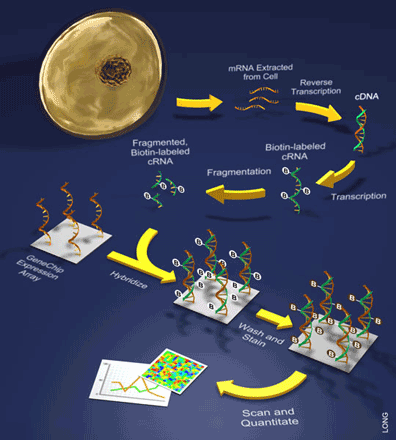
It is important to note that only one sample is hybridized to a single array during these experiments. Such samples are prepared by extracting mRNA from a cell and turning it into back into DNA through a process that involves the reverse transcription of the mRNA into what is referred to as a cDNA (see Figure 2). This is followed by a labeling step during which the cDNA is then transcribed to cRNA while incorporating a label (e.g. biotin). Once labeled, the sample of cRNAs can be hybridized to the array and bound by the various oligonucleotide probes. Lastly, a staining reaction is performed in order to visualize the amount of hybridization.
cDNA Arrays
A cDNA array is a different technology using the same principle; the probes in this case are larger pieces of DNA that are complementary to the genes one is interested in studying [2,5]. Nowadays, cDNA probes for making the array can be generated from a commercially available cDNA library ensuring a close representation of the entire genome of an organism on the array. Alternatively, PCR using specific primers can be used to amplify specific genes from genomic DNA to generate the cDNA probes. A separate PCR reaction must be performed for each gene, although these reactions can be done in parallel. With the cDNA probes prepared they can be mechanically spotted onto a glass slide, normally in duplicate to serve as controls.
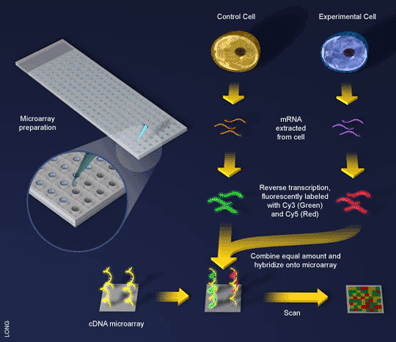
An experiment using a cDNA microarray involves the preparation of two samples for hybridization to the array: a control sample and an experimental sample [5] (see Figure 3). These samples are prepared as before with mRNA being extracted from cells and reverse transcribed into cDNA, with one exception, during the reverse transcription step a fluorescent dye is incorporated into the newly formed cDNA. Here an ingenious trick is used and a different dye employed to label the different samples. For example, the control sample can be labeled with a green-fluorescing dye called Cy3 and the experimental sample labeled with a red-fluorescing dye called Cy5. Since the samples are labeled differently they can be combined and hybridized to the microarray together (see Figure 3). What will happen? The two samples will competitively bind to the probes on the array and the sample containing more gene expression for a particular probe will win out. That is, if there is more of an mRNA transcript in the control sample than in the experimental sample (i.e. the gene is downregulated in the experiment) then more Cy3 will bind to the probe on the array and the spot will fluoresce green. If there is more experimental transcript, the reverse will happen and the spot will fluoresce red. When the two samples have the same amount of transcript, the dyes will cancel each other out and the spot will fluoresce yellow.
When oligonucleotide and cDNA arrays are compared against one another, one will note a few distinctions between them. First, cDNA arrays eliminate the need for the probe design required for oligonucleotide arrays, while also allowing the entire genome of an organism to be represented on the array easily. This makes cDNA arrays more useful for the analysis of gene expression on a global level; for instance, if you wanted to compare the gene expression of an organism when it is grown aerobically versus anaerobically. Whereas, oligonucleotide arrays benefit from their greater hybridization specificity, due to their PM/MM probe design. This, combined with their more specific fluorescence detection (since there are multiple oligonucleotides representing each gene to be studied) make oligonucleotide arrays more useful for monitoring expression levels of a differentially spliced transcript (common in human mRNA). Separate probes for each exon can be printed on the array and heterogeneous transcripts that would all bind to the same cDNA probe will bind differentially to each oligonucleotide probe.
CHIP-ing Away at Today’s Medical Questions
The ability to analyse the activity of every gene in a cell is a powerful tool—it allows us to chip away at the answers to some big questions such as: what is different in the brain of a patient with Alzheimer disease? Can we understand why cancer occurs? Or perhaps, what is happening in a blood vessel that is developing a cardiovascular plaque?
Insights into disease have been one of the most beneficial results of microarray technology [2,6]. For example, the recognition of the loss or reduction of gene expression during a disease can point researchers in important directions. This is because genes that are lost during a disease are typically involved in a cellular function that directly or indirectly prevents the disease from occurring. Identification of these genes with a microarray thus brings about the potential for targeted therapies.
Alternatively, information about differences in gene expression between tissue types can help to uncover how our bodies develop and warn us about which genes would be harmful to target for disease therapy. For example it would not be wise to target a gene that is found to promote cancer but is also important to heart function. This information could then have an important secondary function: disease screening [6-7]. Such genes can be regularly assayed in order to determine a person’s risk of developing a particular disease and even offer insight into the seriousness of disease.
One hotly discussed possibility is that array technology could be used to diagnose diseases at very early stages, so that therapy could be commenced before the disease has the chance to do any harm [6]. Microarrays still need to undergo some optimization before they can be used in such clinical screens, due to the problem of false positive results caused by genes reacting with the array in an inappropriate manner. Current microarray technology at its best still incorrectly report the expression of about 1% of genes it screens. Improvements in the technology are therefore required before such screens are feasible.
Cancer is an especially pertinent target of microarray technology due to the well-known fact that this disease causes, and may even be caused by, changes in gene expression [2]. Microarrays have allowed the rapid identification of which genes are turned on and off in tumour development, resulting in a much better understanding of the disease. For example, if we compare normal cells to cancer cells and discover a gene that is turned on in that particular type of cancer (see Figure 3), we may be able to target that gene for use in cancer therapy. Today, therapies that directly target malfunctioning genes are already in use and showing exceptional results.
Microarrays have many uses in addition to simply identifying the genes that cause a disease to occur [2,6]. Such as, the patterns of correlated loss and increase of gene expression allowing gene interactions to be studied, or the use of microarray analysis in drug design and screening, with compounds that affect the expression of important genes being screened during drug screens [8]. The examples are endless; an indicator of the simple technique’s immense power.
The Expanses of Gene Technology
As molecular biology advances, the technology that supports it advances in ways that were inconceivable a decade earlier. Think of a small piece of glass, reminiscent of a microchip, which is robotically spotted with tens of thousands of gene fragments and then used to scan the unseen molecular events of a cell. An exciting technology to say the least, whose advancement and adaptation should bear heavily on our understanding of biological research.
Additional Reading
1. Sorin D. 2003. Data Analysis for DNA Microarrays. Boca Raton, FL: Chapman & Hall/CRC.
2. Shaw KJ. 2002. Pathogen Genomics: Impact on Human Health. Totowa, NJ: Humana. 309p.
3. Brown SM. 2003. Essentials of Medical Genomics. Hoboken NJ: Wiley-Liss. 274p.
References
1. Southern EM. 2001. DNA Microarrays. History and overview. Methods Mol Biol 170: 1-15.
2. Schena M. 2003. Microarray Analysis. Hoboken, NJ: Wiley-Liss. 630p.
3. Affymetrix GeneChip technology.
4. An overview of how an Affymetrix GeneChip is made.
5. Cheung VG, et al. 1999. Making and Reading Microarrays. Nat Genet 21(1 Suppl): 15-9.
6. Heller MJ. 2002. DNA Microarray Technology: Devices, Systems, and Applications. Annu Rev Biomed Eng 4: 129-53.
7. Jain KK. 2000. Applications of Biochip and Microarray Systems in Pharmacogenomics. Pharmacogenomics 1(3): 289-307.
8. Crowther DJ. 2002. Applications of Microarrays in the Pharmaceutical Industry. Curr Opin Pharmacol 2(5): 551-4.
(Art by Jiang Long – note that high res versions of image files available here)